PYroMat API Documentation
Package structure
The PYroMat package is constructed in two layers, with the user- or command-line layer forming the top and most powerful layer. The base PYroMat modules provides a minimalist interface to the more sophisticated back-end.
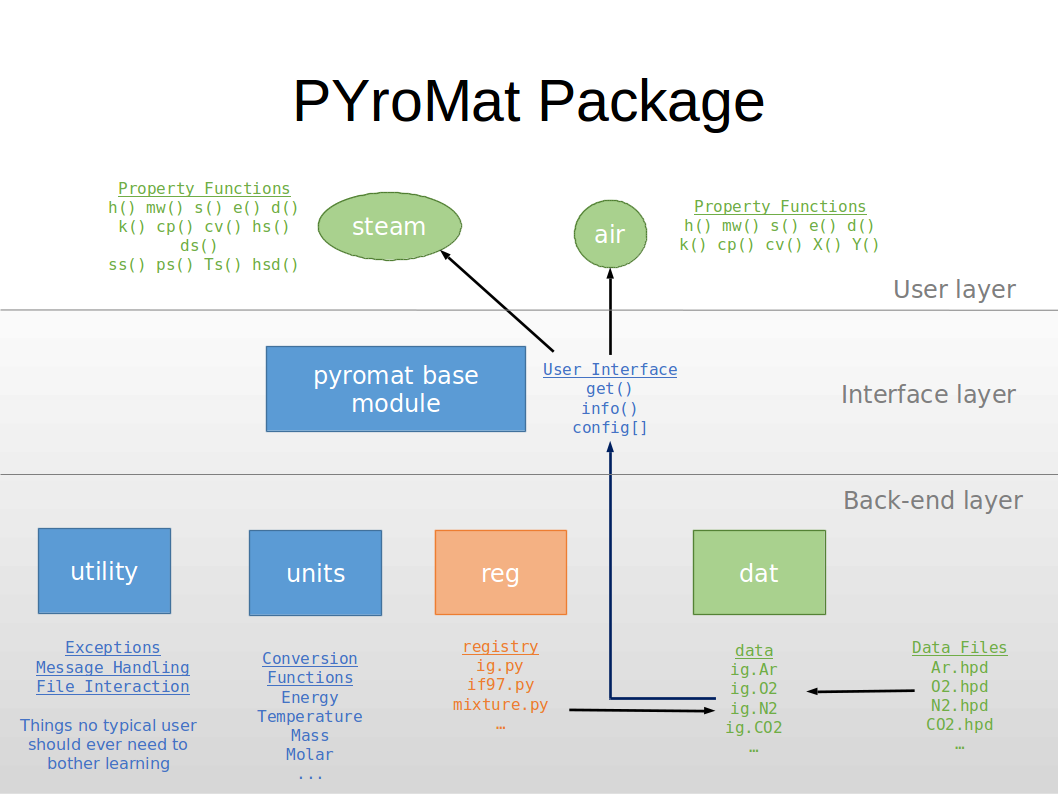 Figure 1: The PYroMat package structure
Figure 1: The PYroMat package structure
When a user calls for an object like "ig.N2" (see the tutorial), the get() function reaches into the dat module to see if PYroMat has any entries that match the ID the user requested.
All of the jobs needed to get the package to the point where such a thing is possible are parsed out to the various back-end layer modules. The dat module is responsible for seeking out data files, loading them, and associating them with the correct data object in its data dictionary.
The reg module is responsible for seeking out python files that might contain PYroMat object class definitions. Candidate files are compiled and executed in their own name space. Any class definitions that are children of the PYroMat __basedata__ prototype class are added to the registry dictionary where the dat module can find them.
The units module's purpose is pretty self-explanatory, but its implementation is not. It holds a collection of funcitons that are named for the types of unit conversions they can perform. These functions are made special because they are aware of PYroMat's unit configuration options. Users who want to see what options are available should call up the units.show() function. This module also has a collection of useful constants like the Boltzmann constant (const_k), the universal gas constant (const_Ru), and others.
Finally, the utility module is a catch-all for every function, exception, class, and other miscelania that the package needs to operate. That includes the configuration class on which the config object is built, all exceptions, and file handling functions for loading, testing, and maintaining the data files.
TopInterface layer
The vast majority of functionality a typical user will need is provided in the user layer by the individual data objects. The interface layer is only responsible for querying and retrieving those objects.
get() and info()
For the time being, there are only two functions that are used for interacting with the PYroMat data objects; get() and info(). When called with no arguments, the info() function prints a table of the IDs for all the available data objects. If called with the ID string of one of those objects, the info() function prints documentation for the object. Usually this will include a description of the object and citations for the data source.
By far the most important member of the interface layer, the get() function retrieves the data objects. Really, a call to get(idstring) is quite similar to pyromat.dat.data[idstring], but with some graceful error handling.
config[]
The configuration object, config, manages all of the user-configurable behaviors of PYroMat. That includes how data are loaded, which units should be used, and what temperature-pressure defaults should be used. The config object is an instance of the PMConfig class and its entries are all PMConfigEntry instances; both classes provided in utility module.
The config object is a wrapper for a dictionary, but with algorithms for building itself from user-written configuration files with built-in default values and with rules on acceptable values for each entry. Entries remember whether they are read-only, they always remember their default values, and they raise exceptions if they are written with illegal values. A complete list of the available configuration options and their current values is obtained simply be evoking the config object at the command line.
>>> pm.config
config_file : ['/home/chris/Documents/PYroMat/pyromat/defaults.py']
config_verbose : False
dat_dir : ['/home/chris/Documents/PYroMat/pyromat/data']
dat_exist_fatal : False
dat_overwrite : True
dat_recursive : True
dat_verbose : False
def_T : 298.15
def_p : 1.01325
install_dir : '/home/chris/Documents/PYroMat/pyromat'
reg_dir : ['/home/chris/Documents/PYroMat/pyromat/registry']
reg_exist_fatal : False
reg_overwrite : True
reg_verbose : False
unit_energy : 'kJ'
unit_force : 'N'
unit_length : 'm'
unit_mass : 'kg'
unit_matter : 'kg'
unit_molar : 'kmol'
unit_pressure : 'bar'
unit_temperature : 'K'
unit_time : 's'
unit_volume : 'm3'
version : '2.0.1'
The configuration entries are organized into several groups based on how they are used. For example, entries beginning with reg_ and dat_ determine how class definitions and data files are loaded. Entries beginning with unit_ specify the default units to be used by the units module. def_T and def_p indicate the default temperature and pressure to be used by property methods when the arguments are omitted.
Some entries are read-only.
>>> import pyromat as pm >>> pm.config['version'] '2.0.1' >>> pm.config['version'] = 42 PYroMat ERR:: Failed to write to configuration parameter, 'version' (, PMParamError('Entry is read-only.',), ) Traceback (most recent call last): File " ", line 1, in File "pyromat/utility.py", line 347, in __setitem__ raise sys.exc_info()[1] pyromat.utility.PMParamError: Entry is read-only.
Entries are data-type specific.
>>> pm.config['def_T'] 298.15 >>> pm.config['def_T'] = 300. >>> pm.config['def_T'] 300.0 >>> pm.config['def_T'] = 'This is NOT a temperature' PYroMat ERR:: Failed to write to configuration parameter, 'def_T' (, PMParamError("Expected , but got 'This is NOT a temperature'",), ) Traceback (most recent call last): File " ", line 1, in File "pyromat/utility.py", line 347, in __setitem__ raise sys.exc_info()[1] pyromat.utility.PMParamError: Expected , but got 'This is NOT a temperature'
And some entries behave a little strangely when they're written to. Instead of overwriting the old value, new values are appended to a list. These are usually lists of directories or files where PYroMat should search for data. They can be reset to safe defaults using the restore_default() method.
>>> pm.config['config_file']
['/home/chris/Documents/PYroMat/pyromat/defaults.py']
>>> pm.config['config_file'] = '/etc/pyromat/pyromat.py'
>>> pm.config['config_file']
['/home/chris/Documents/PYroMat/pyromat/defaults.py', '/etc/pyromat/pyromat.py']
>>> pm.config.restore_default('config_file')
>>> pm.config['config_file']
['/home/chris/Documents/PYroMat/pyromat/defaults.py']
Changes that are made in scripts and at the command line are lost when a user exists Python. To make changes persistent, they should be placed in a configuration file. The configuration process is described in detail below.
TopThe import process
When PYroMat is imported for the first time, the package goes through a series of steps to build up its capabilities from nothing. At its core, PYroMat doesn't know the first thing about substances of any kind. For each substance, there is a file somewhere that provides all the necessary property data and names a class in the PYroMat "registry" with the methods for interpreting the data. In the same way, there is a file somewhere that contains the definition for each class in the registry.
Where do those files exist? In what order should they be loaded? What happens if there are redundant definitions? How do users add their own files without having administrator priviledges? All of these questions and more are addressed by the PYroMat configuration system.
Loading the configuration
The whole process begins with loading the system's configuration files. Just like lots of other configuration systems, PYroMat uses parameter-value pairs. Instead of using some other syntax, PYroMat configuration files are just Python scripts. All variables defined in the local name space are written to the configuration object, config. Any variables that are not recognized PYroMat configuration directives will result in an error, and so will illegal values.
When PYroMat begins its own import process, it is aware of a single configuration file, "defaults.py," in the PYroMat installation directory. It is always loaded first, and it allows administrators to specify additional places to look for configuration files (like in users' home directories).
The config_file directive is one of several that append new values to a list rather than overwriting the old value, so each time a configuration file contains a config_file directive, the list of files grows. Even though the initial list contained only a single file, the configuration's load() method continues recursively until the growing list is finally exhausted. The savvy reader might worry about accidentally creating an endless cyclic references. Good for you! The configuration system maintains a list of all the files it has loaded and checks each file against the list. No file will be loaded twice.
For example, on a Unix system, an administrator might want to set up a configuration script in /etc. This line should be added to the "defaults.py" file: config_file = '/etc/pyromat.py'. Then, to allow users to write their own configurations (maybe you have some people who like different unit systems), that file might look something like this:
# PYroMat configuration file #/etc/pyromat.py # Multiple entries should appear in a list # Note that environment variables work config_file = ['/home/$USER/.pyromat/config.py', '/home/$USER/.pyromat.py'] # Note that the ~ symbol in Unix is also supported dat_dir = '~/.pyromat/data' # Of course, explicit definitions work too reg_dir = '/usr/share/pyromat/reg'
It is important to note that if a directive is overwritten within the same script, then the rules about appending values will not be applied. These are only used to join the values found in the different configuration files. In the above example, config_file is written with a list value in order to add two entries at once.
For detailed documentation on all the configuration directives, read the comments in the "defaults.py" file.
Populating the registry
The registry is where all the class definitions reside. The reg module has a function called regload() that is responsible for seeking out the contents of all the directories identified by the reg_dir configuration directive. Like the config_file directive, it accumulates values from all configuration files.
Unlike configuration files, class definition files do not need to be called out by name. Instead, they only need to be placed in one of the directories named in reg_dir. The file name must have a .py extension, and all files with an underscore "_" or a dot "." prefix in their name are ignored. Files that meet these criteria are compiled and executed. All objects that are type instances and are subclasses of the reg.__base_data__ class are incorporated into the reg.registry dictionary.
In this documentation, "the registry" refers to that dictionary of classes. They keys to the dictionary are the names of the classes they define. This is important because the class name is called out by each data file.
There are three True/False-valued configuration options that affect the behavior of the registry: reg_verbose, reg_overwrite, and reg_exist_fatal. When set to True reg_verbose prompts reg.regload() to print its behavior to stdout, which can be helpful for debugging.
The latter two deal with file precedence in the case of a redundant definition. When True, reg_overwrite a class definition that is named identically with one already loaded will replace its predecessor. Otherwise, the first loaded receives preference. If there is a redundant definition when reg_exist_fatal is True, the load process will fail with a PMFileError exception.
For multiple reasons, the registry construction process never recurses into sub-directories. First of all, registries do not usually require many files, and permitting complicated directory structures is not really necessary. The real reason recursion is not supported is to force users to be extremely deliberate about the codes they intend to include in the registry.
Valid PYroMat data classes must have initializers that accept a data dictionary as a single argument. What the class chooses to do with those data are up to it, but they are passed on from the data file load process.
Populating the data dictionary
Just like the registry load process populates the registry with class definitions, the data load process populates a dictionary, dat.data with all the individual species objects defined in the PYroMat system.
Each data file defines the properties of a single species in a JavaScript Object Notation (JSON) format. All data files must have the extension *.hpd, which is a throwback to the package's 2014 working title: Hot Python Data. While the content and meaning of the individual data elements are entirely up to the class, PYroMat requires several elements to be defined in the resulting dictionary.
Each data file must define a value, id, which is the key that will be used to recognize the data object in the data dictionary. Each file must have a string class value, which will be used to call an element of reg.registry to create the data object. Finally, each file must define doc, which is a long string used to supply a description of the species source data. After loading, each data dictionary is forced to have the fromfile key, which indicates the absolute path to the original data file.
While the actual data loading algorithm is more complicated, adding a data file looks something like this:
import json
import pyromat as pm
with open("mydata.hpd") as ff:
new_data_dict = json.load(ff)
new_data_class = pm.reg.registry[new_data_dict['class']]
pm.dat.data[new_data_dict['id']] = new_data_class(new_data_dict)
For developers, the dat.load() function has a number of additional documented features that can help with debugging. In particular, files can be excluded by adding a tilde "~" after the *.hpd extension. PYroMat will regard this as a "suppressed" data file; one that can be but is not currently included in the load process.
The data loading process is controlled by four configuration parameters. Like the registry load process, dat.load() can be prompted to print its behavior to stdout when the dat_verbose directive is set to True. Similarly, there are dat_overwrite and dat_exist_fatal options to control precedence in the case of redundant definitions. Unlike the registry, data file searches will recurse into subdirectories when the dat_recursive directive is set to True.
Once this process is complete, the package is fully functional. The get() function is little more than a wrapper for the dat.data dictionary.
TopBack-end layer
reg
The reg module is responsible for maintaining the registry; a dictionary of PYroMat class definitions that provide users all the tools they need to interact with the species' data. The reg module provides three essential tools:
- The regload() function
- The registry dictionary
- The __basedata__ class definition
The regload() function is responsible for loading the basic class definitions used to spawn all the individual species' objects described in the import process. The function's behavior is determined by the configuration options reg_dir, reg_overwrite, reg_exist_fatal, and reg_verbose. Their behaviors are discussed with the import system. Each file found with a ".py" extension is compiled and executed in its own variable space, and any objects that are children of the __basedata__ class will be added to the registry.
The registry is the dictionary that contains all of the class definitions found by regload(). The key is the string __name__ of the class definition, and the value of each is the class object itself.
The __basedata__ class must be a parent of all PYroMat class definitions. It defines an initializer and a __basetest__() member that does some VERY simple integrity checking on the data. In earlier versions, __basedata__ also offered some tools for "vectorizing" data and a psolve routine for property inversion, but these were depreciated in favor of other methods. The most important member of __basedata__ is data, which is the dictionary which is supposed to contain the species' data.
By default, each class definition resides in its own appropriately named file in the "registry" directory. Users who want to develop their own classes might want to change that by adding entries to the reg_dir configuration directive. Here is a minimalist custom class definition that references the __basedata__ class, and calls its __init__ method to do some things we will discuss in the next section.
import pyromat as pm
class MyClass(pm.reg.__baseclass__):
def __init__(self, data):
pm.reg.__baseclass__.__init__(self,data)
dat
The dat module is responsible for maintaining the data dictionary, which provides each of the individual species objects keyed by their species ID. In addition to being able to load these objects, the dat module offers tools to create, manipulate, and even save them. In addition to the data dictionary itself, the dat module contains:
| load() | Loads the PYroMat ".hpd" files into the data dictionary. If called with the check=True option, instead of populating data, load() will load files into a fresh dictionary, and perform a detailed comparison with the data currently in memory. |
| clear() | Empties the data dictionary. |
| new() | Creates a new entry in the data dictionary from a dictionary of raw data. The dictionary must contain "id", "class", and "doc" keys, so that new() will know how to store the object and with which class to associate it. Otherwise, it must only conform to the rules of the class. |
| updatefiles() | When run verbosely, this function inters an interactive dialogue to bring the files into agreement with the data currently in memory. The idea is that users will be free to edit entries in the data dictionary directly and the files can be automatically updated to make the changes permanent. |
More detail is offered by the in-line help on each of these functions.
When load works its way through the dat_dir list of directories looking for files with the ".hpd" extension, it uses the json module to load them. The result is a dictionary with entries for each of the data elements defined in the file. This dictionary is passed verbatim to the class initializer for each object, so that the creation of the oxygen object might appear:
>>> import pyromat as pm
>>> import json
>>> with open('O2.hpd','r') as ff:
... data = json.load(ff)
...
>>> class = data['class']
>>> O2 = pm.reg.registry[class](data)
The behavior of load() is defined by configuration directives, dat_dir, dat_overwrite, dat_exist_fatal, and dat_verbose. These are described in more detail in the import system section.
units
The unit conversion system uses a single class as the prototype for a collection of function-like objects that go between the various unit systems PYroMat recognizes. The fastest way to see what is available is to call the show() function.
>>> pm.units.show()
force : lb lbf kN N oz kgf
energy : BTU kJ J cal eV kcal BTU_ISO
temperature : K R eV C F
pressure : mmHg psi inHg MPa inH2O kPa Pa bar atm GPa torr mmH2O ksi
molar : Ncum NL Nm3 kmol scf n mol sci Ncc lbmol
volume : cumm cc mL L mm3 in3 gal UKgal cuin ft3 cuft USgal m3 cum
length : ft nm cm mm m km um mile in
mass : mg kg g oz lb lbm slug
time : s ms min hr ns year day us
Each row is named for the Conversion object that defines the units listed. For example, in order to convert a force from 4.5 newtons to pounds,
>>> pm.units.force(4.5, from_units='N', to_units='lb') 1.0116402439486973
When the from_units or to_units keywords are omitted, then each of the Conversion objects is linked to a configuration directive to use instead. This is how the various PYroMat data classes convert to and from the user-specified unit systems. For example,
>>> pm.config['unit_force'] = 'N' >>> pm.units.force(4.5, to_units='lb') 1.0116402439486973 >>> pm.units.force(1.0116402439486973, from_units='lb') 4.5
Of course, some conversions require multiple steps. For example, molecular weight is specified in unit_mass per unit_molar. To make that multi-step process easier, there is an optional exponent keyword to indicate the unit's exponent in the unit cluster.
>>> O2mw = 32. # kg / kmol >>> O2mw = pm.units.mass(O2mw, from_units='kg', to_units='lbm') >>> O2mw = pm.units.molar(O2mw, from_units='kmol', to_units='mol', exponent=-1)
This calculates molecular weight in pound-mass per mole; if you like that kind of thing.
This behavior works for all nine (9) of the Conversion instances above, but there are also two special cases: temperature_scale() and matter().
The temperature() conversion tool will perform temperature conversions with the assumption that all temperatures are
>>> pm.units.temperature(32., from_units="F", to_units="C") 17.77777777777778 >>> pm.units.temperature_scale(32., from_units="F", to_units="C") 0.0
The matter() function is a different matter. It requires the molecular weight of a substance, and it can convert between mass and molar units. This is what PYroMat uses to handle intensive units like entropy, enthalpy, specific heat, etc. It is linked to the unit_matter configuration directive for its default behavior.
>>> pm.units.matter(1., mw=32., from_units="kmol", to_units="kg") 32.0
The units module also exposes a number of important constants on which it depends for its calculations.
| const_k | Joule/Kelvin | Boltzmann's constant |
| const_Na | count/mole | Avagadro's constant |
| const_Ru | Joule/mole/Kelvin | Universal gas constant |
| const_Tstd | Kelvin | Standard temperature for volumetric molar units |
| const_pstd | bar | Standard pressure for volumetric molar units |
| const_Nstd | mole/cubic meter | Standard number density for volumetric molar units |
| const_g | meter/second2 | Acceleration due to gravity |
| const_dh2o | kg/meter3 | Density of water used for column pressure |
| const_dhg | kg/meter3 | Density of mercury used for column pressure |
These and all of their dependent Conversion objects are initialized using the setup() function when the module is first loaded. To change any of these constants, see the setup() in-line documentation. Evoking it at the command line will re-define all of the dependent routines correctly.
utility
Coming soon. Move along, there's nothing to see here. These aren't the documents you're looking for. The cake is a lie. Seriously, why do you want to read this anyway; I'm sure it would be really boring. OK fine. I'll get to it later.
TopIdeal gas collection (ig)
The properties of all members of the ideal gas collection conform to several important assumptions:
- Specific heats (and therefore enthalpy) is not a function of pressure.
- Density is computed from temperature and pressure by the ideal gas equation (i.e. compressibility is 1).
- Isotopes of the substance can be neglected when calculating molecular weight.
For properties like enthalpy and entropy that are defined in terms of integrals of specific heat, there is an arbitrary choice of integration constant. These are usually selected so that a property will have a convenient value at a convenient state. However, because of their importance to reaction modeling, these ideal gas properties are all standardized to permit the calculation of properties across chemical reactions.
All members of the ideal gas collection support an optional True/False parameter, hf, to the enthalpy method, which allows users to disable the inclusion of the "enthalpy of formation." When hf=False, the enthalpy of the species will be calculated so that h(298.15, 1.) is quite close to zero (in K and bar). When hf=True (the default) the enthalpy will include the energy released/consumed due to chemical reactions, and only a few so-called "reference species" will have zero enthalpy at standard conditions (i.e. N2, O2, H2, Ar, etc.).
All arguments and all returned values are translated according to the PYroMat unit configuration directives. The in-line documentation for each method specifies which units directives are used to convert inputs and outputs. It should be emphasized that specifying unit mass, unit length and unit time does NOT specify the unit energy.
Ideal gas (ig)
The ideal gas class is built on the so-called Shomate equation of state, which is used both by the NIST Webbook and Cantera to calculate ideal gas properties. It is a piece-wise five-term expansion for isobaric specific heat with a single non-polynomial term. The ideal gas class provides properties:
| cp(T,p) | isobar specific heat | unit_energy / unit_temperature / unit_matter |
| cv(T,p) | isochor specific heat | unit_energy / unit_temperature / unit_matter |
| d(T,p) | density | unit_matter / unit_volume |
| e(T,p) | internal energy | unit_energy / unit_matter |
| h(T,p) | enthalpy | unit_energy / unit_matter |
| gam(T,p) | specific heat ratio | dimensionless |
| mw() | molecular weight | unit_mass / unit_molar |
| R() | gas constant | unit_energy / unit_temperature / unit_matter |
| s(T,p) | entropy | unit_energy / unit_temperature / unit_matter |
| T_h(h,p) | temperature from enthalpy |
| T_d(d,p) | temperature from density and pressure |
| T_s(s,p) | temperature from entropy and pressure |
| p_s(s,T) | pressure from entropy and temperature |
| p_d(d,T) | pressure from density and temperature |
The method, Tlim(), returns a two-element tuple with the min,max temperatures supported by the data set.
These data are primarily useful at high temperatures, but the exact limits on the data are not consistent across species. Some are limited by phase transitions, and others simply by the source data. Each object reports its own limits using the Tlim() method. Calls to properties outside of these limits will result in an error.
Finally, the contents() method returns a dictionary that names the constituent atoms and their quantities in the species chemical formula. It does this by parsing the species ID for upper-case letters followed by lower-case letters, followed by an optional integer quantity. The result might look like {'C': 1, 'O': 2} for carbon dioxide.
Ideal gas mixture (igmix)
The ideal gas mixture class permits the calculation of properties of mixtures of ideal gases. These data belong to the ideal gas collection because they follow all of the same rules, rest on the same physical assumptions, and are built on the same base data. The igmix methods work by combining the relevant member species' ig methods in the appropriate weighted averages.
The valid temperature range for ideal gas mixtures is limited by the range of which ALL constituents have valid data. Some ideal gases (like diatomic oxygen) support low temperatures, but most do not. As a result, mixtures that contain oxygen (like air) may not support the same broad range as each constituent. For that reason, igmix has its own Tlim() method that reports the intersection of all the constituent species' temperature ranges.
The igmix class provides properties:
| cp(T,p) | isobar specific heat | unit_energy / unit_temperature / unit_matter |
| cv(T,p) | isochor specific heat | unit_energy / unit_temperature / unit_matter |
| d(T,p) | density | unit_matter / unit_volume |
| e(T,p) | internal energy | unit_energy / unit_matter |
| h(T,p) | enthalpy | unit_energy / unit_matter |
| gam(T,p) | specific heat ratio | dimensionless |
| mw() | molecular weight | unit_mass / unit_molar |
| R() | gas constant | unit_energy / unit_temperature / unit_matter |
| s(T,p) | entropy | unit_energy / unit_temperature / unit_matter |
| X() | mole fractions | dimensionless |
| Y() | mass fractions | dimensionless |
The igmix class provides inverse property routines:
| T_h(h,p) | temperature from enthalpy |
| T_s(s,p) | temperature from entropy and pressure |
Multi-phase collection (mp)
The multi-phase collection is home to classes that support the calculation of species' properties across phase changes. Unlike the ideal gas collection, the integration constants in these species do not include the enthalpies of formation.
Steam class (if97)
The International Association for the Properties of Water and Steam issued the Industrial Formulation for water's equation of state in 1997. The report is abbreviated IAPWS-IF97, or IF97 for short, and has been widely adopted for applied use because it permits direct calculation of properties from temperature and pressure. Many precise two-phase equations of state use temperature and density to provide an elegant mathematical description of the phase change (which T,p does not permit). However, in applications where temperature and pressure are known, this requires expensive iteration to invert the formulation.
The if97 class implements the piece-wise polynomial fit for water and steam. In addition to the basic properties like specific heats, enthalpy, entropy, and internal energy, if97 also includes special methods for evaluating properties along the saturation line. Almost all of the properties accept or return quality, x, as an optional parameter. See the in-line documentation for a detailed description of each method.
The if97 class provides the following properties:
| cp(T,p) | isobar specific heat | unit_energy / unit_temperature / unit_matter |
| cv(T,p) | isochor specific heat | unit_energy / unit_temperature / unit_matter |
| d(T,p) | density | unit_matter / unit_volume |
| ds(T,p) | saturation density | unit_matter / unit_volume |
| e(T,p) | internal energy | unit_energy / unit_matter |
| es(T,p) | saturation energy | unit_energy / unit_matter |
| h(T,p) | enthalpy | unit_energy / unit_matter |
| hs(T,p) | saturation enthalpy | unit_energy / unit_matter |
| hsd(T,p) | enthalpy, entropy, density | (see h,s, and d |
| gam(T,p) | specific heat ratio | dimensionless |
| mw() | molecular weight | unit_mass / unit_molar |
| s(T,p) | entropy | unit_energy / unit_temperature / unit_matter |
| ss(T,p) | saturation entropy | unit_energy / unit_temperature / unit_matter |
| Ts(p) | saturation temperature | unit_temperature |
| ps(T) | saturation pressure | unit_pressure |
| triple() | triple point (T,p) | unit_temperature, unit_pressure |
| critical() | critical point (T,p) | unit_temperature, unit_pressure |
The if97 provides inverse routines:
| T_h(h,p) | temperature from enthalpy |
| T_s(s,p) | temperature from entropy and pressure |
The limits on the validity of the IF-97 class properties are reported by the plim() and Tlim() methods. Since the IF-97 report equations are valid over piece-wise rectangles in T,p space, these functions accept temperature and pressure respectively as optional arguments.
Top